Simulation of Micellar Self-Formation using LAMMPS
Last Update:2021/12/09
1. Introduction
The molecular dynamics software LAMMPS can be used to handle soft matter problems. In particular, it is relatively easy to create impressive movies by simulating and visualizing the formation of structures such as membranes. Let’s take the example of micelle in the LAMMPS package and visualize it with the visualization software ovito. The operating environment is MateriApps LIVE! version 3.3.
2. Preparation
First, create a working directory in your home directory and move to the working directory.
cd $HOME mkdir lammps cd lammps
Then copy the LAMMPS examples/micelle directory and move to it.
cp -r /usr/share/lammps/examples/micelle ./ cd micelle
3. Editing an input file
You will find the input file in.micelle in the micelle directory. Edit this file to visualize the state during the calculation.
emacs in.micelle
Any editing software will do. Emacs is used here, but MateriApps LIVE! includes other editors such as vim, nano, and Mousepad, so choose your favorite editor. The contents of the file are as follows:
# 2d micelle simulation dimension 2 neighbor 0.3 bin neigh_modify delay 5 atom_style bond # Soft potential push-off read_data data.micelle special_bonds fene ... thermo 50 #dump 1 all atom 2000 dump.micelle ... reset_timestep 0 run 1000
In the beginning, dimension 2 specifies a two-dimensional calculation. At the last part of the file, on the line starting with “#dump 1 …”, omit the first “#” and change this line as
dump 1 all atom 2000 dump.micelle
In addition, the last line of the input file “run 1000” is modified as
run 100000
After finishing the edit of the input file, save the file and exit the editor. (In Emacs, save with ctrl-x ctrl-s and exit with ctrl-x ctrl-c.)
4. Performing Calculations with LAMMPS
Run LAMMPS using the edited input file in.micelle.
lammps < in.micelle
If there is no error message and “Total wall time” is displayed at the end, the execution has completed successfully. The file dump.micelle is now generated in the same directory. (You can check it with the ls command.)
5. Visualization by ovito
Open the generated file with ovito.
ovito dump.micelle
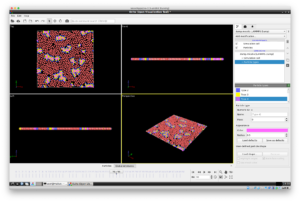
You should see the following screenshot (This screeenshot is after pressing the play button, but there should be a screen where micelles are not formed yet). Press the button at the bottom right to play the video. You can see the particles gathering to form a micellar structure.
6. Summary
This is an easy visualization for relatively small molecular dynamics calculations. For visualization of systems containing many particles, the visualization becomes heavy in the environment of MateriApps LIVE!. Therefore, it is more efficient to install visualization software (such as VMD) directly on your desktop and visualize it after transferring the dump data to your machine.
