Molecular dynamics simulation of a peptide chain using Gromacs
Last Update:2021/12/09
Introduction
Gromacs is open source software that performs molecular dynamics simulations. It has a number of useful functions for dynamics calculation of biomolecules such as proteins and nucleic acids, and is often used in the fields of biology and pharmacy. Structure data of nucleic acids and proteins can be downloaded from a database (such as PDB). Based on the structural data of these biomolecules, molecular dynamics calculations are performed after solvent molecules such as water are arranged. Molecular dynamics simulations provides useful information, which is used, e.g., in the analysis of interactions between proteins and ligands. Here, let’s perform molecular dynamics calculations of peptide chains in water on MateriApps LIVE!.
The version of MateriApps LIVE! Used in this tutorial is 2.4.
How to run
Using a script file prepared in advance, we simulate the dynamics of a small peptide chain called [KIGAKI]3 in water (for details, see GromacsTutorial). Since the calculation time is long, it is better to increase the number of CPUs as much as possible in the setting of VirtualBox in advance. You can change the number of CPU processors in VirtualBox by going to “Settings>System>Processors”. (Check the number of the CPU cores of the PC. Normally, the user should set the maximum within the range of the green bar on the CPU number selection screen.) Next, start MateriApps LIVE!, log in, select “System Tools> LXTerminal” from the start menu on the lower left of the screen, and start up the terminal. Type the following commands in a terminal and press the return key after each line:
$ wget https://github.com/cmsi/malive-tutorial/releases/download/em2020/gromacs.tgz $ tar zxvf gromacs.tgz $ cd gromacs $ sh run.sh
The degree of parallelism is automatically determined by the software side. This molecular dynamics calculation should be completed in about 3 to 5 hours on a notebook PC and about 1 hour on a high-performance desktop machine.
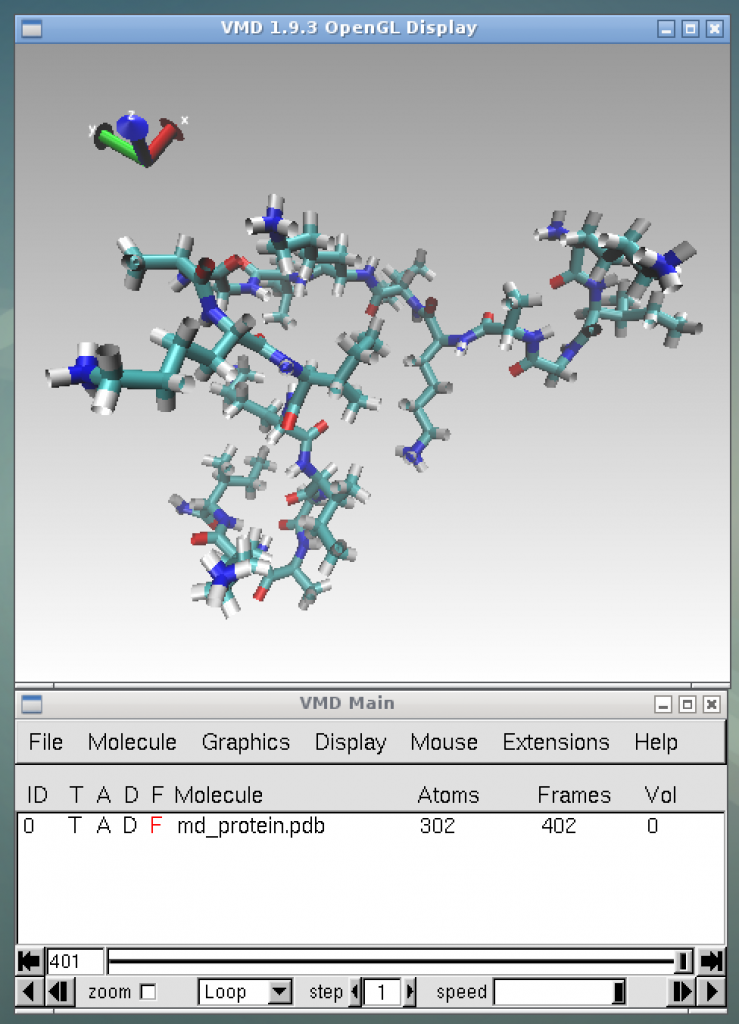
For visualization, we use VMD. MateriApps LIVE! provides only a GUI-based setup tool (VMD Setup) due to the license problem. First, Click “Download VMD”in the VMD web page and select the appropriate version and OS. Register the user name and password. (File download may start, but this file is not necessary.) Next, select “Start menu > MateriApps > VMD Setup” on MaterApps LIVE!, enter the registered user name and password in the window that appears, and click the “Install” button. After VMD is installed, execute the following commands from the terminal:
$ sh animation.sh
This command will visualize the molecular structure of [KIGAKI]3 as shown in the picture below. If the user press the play button at the bottom right of the bottom window, the animation of the time evolution of the molecular structure will start.
Setting up Gromacs is a little complicated. You can see that various commands are described in the script file as you can see it by typing “more run.sh” in the terminal and pressing the return key. In these commands, a simulation box is created and water molecules are located. The original molecular structure file is given by a PDB file (extension: .pdb. For details, see GromacsTutorial). The calculation conditions are given in the .mdp file.
Summary
The selection of the force field (which is selected by the first command in the “run.sh” script) is important in Gromacs. The protein structure file (PDB file) can be downloaded from PDB, but some small structures and hydrogen atoms may be omitted in the date, and often the user needs to complement them. Of course, for large molecules such as proteins, computation requires a certain amount of computational resources, and MateriApps LIVE!, which uses a virtual environment, is not powerful enough. However, if you install Gromacs directly on a PC or computer server with some specifications, molecular dynamics simulation can be completed in a matter of hours to a day. Now you can perform protein dynamics calculations, which were once only handled by supercomputers, on your PC!